You need to agree to share your contact information to access this model
This repository is publicly accessible, but you have to accept the conditions to access its files and content.
You agree to not use the model to conduct experiments that cause harm to human subjects.
Log in or Sign Up to review the conditions and access this model content.

COntrastive Biomarker Representation Alignment (COBRA)
Abstract
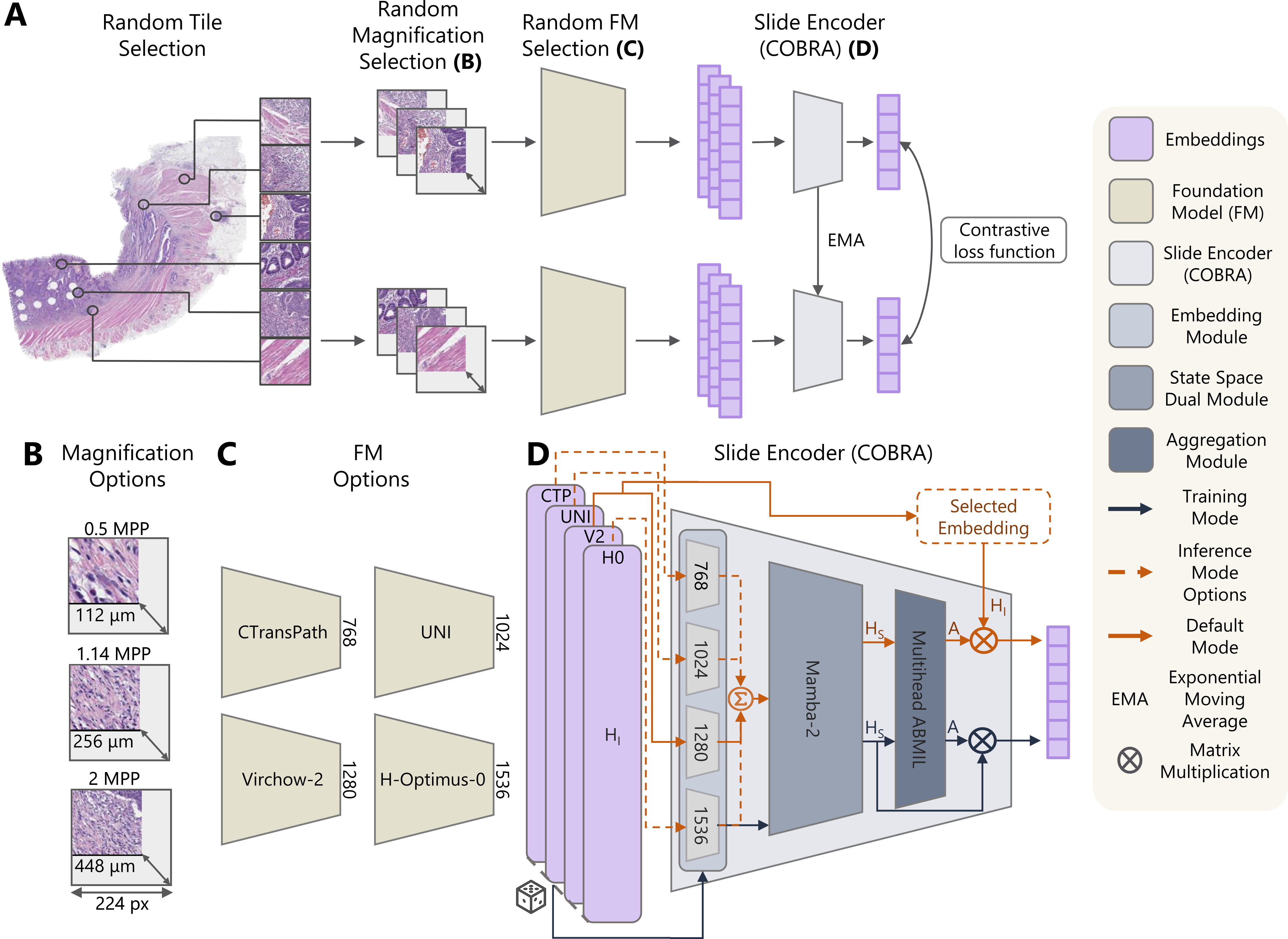
Representation learning of pathology whole-slide images(WSIs) has primarily relied on weak supervision with Multiple Instance Learning (MIL). This approach leads to slide representations highly tailored to a specific clinical task. Self-supervised learning (SSL) has been successfully applied to train histopathology foundation models (FMs) for patch embedding generation. However, generating patient or slide level embeddings remains challenging. Existing approaches for slide representation learning extend the principles of SSL from patch level learning to entire slides by aligning different augmentations of the slide or by utilizing multimodal data. By integrating tile embeddings from multiple FMs, we propose a new single modality SSL method in feature space that generates useful slide representations. Our contrastive pretraining strategy, called COBRA, employs multiple FMs and an architecture based on Mamba-2. COBRA exceeds performance of state-of-the-art slide encoders on four different public Clinical Protemic Tumor Analysis Consortium (CPTAC) cohorts on average by at least +4.5% AUC, despite only being pretrained on 3048 WSIs from The Cancer Genome Atlas (TCGA). Additionally, COBRA is readily compatible at inference time with previously unseen feature extractors. Code available at https://github.com/KatherLab/COBRA.
News
- [Feb 7th 2025]: COBRA II trained on all TCGA cohorts, is now live and ready to use!!
Installation
To install the necessary dependencies, run the following commands:
git clone https://github.com/KatherLab/COBRA.git && cd COBRA
pip install uv
uv venv --python=3.11
source .venv/bin/activate
uv pip install "torch==2.4.1" setuptools packaging wheel "numpy==2.0.0"
uv sync --no-build-isolation
WSI Level Embeddings
To deploy the COBRA model, follow these steps:
- Prepare your data: extract tile embeddings with one or more patch encoders of your preference using STAMP.
- COBRA I:
- supported tissue types: LUAD, LUSC, STAD, CRC, BRCA
- supported patch encoders to generate weighting:
- CTransPath, UNI, Virchow2, H_optimus_0
- supported patch encoders for patch feature aggregation:
- all existing patch encoders compatible with patch size 224x224 px
- COBRA II:
- supported tissue types: all cohorts included in TCGA
- supported patch encoders to generate COBRAII weighting:
- CONCH, UNI, Virchow2, H_optimus_0
- supported patch encoders for patch feature aggregation with COBRAII:
- all existing patch encoders compatible with patch size 224x224 px
- COBRA I:
- Request Access.
- Deploy COBRA:
- extract slide level embeddings using COBRAI/II
python -m cobra.inference.extract_feats_slide --feat_dir <tile_emb_dir> --output_dir <slide_emb_dir> (--checkpoint_path <checkpoint_path> --config <path_to_config> | -d)
- extract patient level embeddings using COBRAI/II
python -m cobra.inference.extract_feats_patient --feat_dir <tile_emb_dir> --output_dir <slide_emb_dir> --slide_table <slide_table_path> (--checkpoint_path <checkpoint_path> --config <path_to_config> | -d)
References
- CTransPath
Xiyue Wang, Sen Yang, Jun Zhang, Minghui Wang, Jing Zhang, Wei Yang, Junzhou Huang, and Xiao Han. Transformer-based unsupervised contrastive learning for histopathological image classification. Medical Image Anal- ysis, 2022
- UNI
Richard J Chen, Tong Ding, Ming Y Lu, Drew FK Williamson, Guillaume Jaume, Bowen Chen, Andrew Zhang, Daniel Shao, Andrew H Song, Muhammad Shaban, et al. Towards a general-purpose foundation model for com- putational pathology. Nature Medicine, 2024
- Virchow2
Eric Zimmermann, Eugene Vorontsov, Julian Viret, Adam Casson, Michal Zelechowski, George Shaikovski, Neil Tenenholtz, James Hall, David Klimstra, Razik Yousfi, Thomas Fuchs, Nicolo Fusi, Siqi Liu, and Kristen Sever- son. Virchow2: Scaling self-supervised mixed magnification models in pathology, 2024
- H-Optimus-0
Charlie Saillard, Rodolphe Jenatton, Felipe Llinares-López, Zelda Mariet, David Cahané, Eric Durand, and Jean-Philippe Vert. H-optimus-0, 2024
- CONCH
Lu, Ming Y. and Chen, Bowen and Zhang, Andrew and Williamson, Drew F. K. and Chen, Richard J. and Ding, Tong and Le, Long Phi and Chuang, Yung-Sung and Mahmood, Faisal. A visual-language foundation model for computational pathology. Nature Medicine, 2024
- STAMP
Omar S. M. El Nahhas, Marko van Treeck, Georg Wölflein, Michaela Unger, Marta Ligero, Tim Lenz, Sophia J. Wagner, Katherine J. Hewitt, Firas Khader, Sebastian Foersch, Daniel Truhn, and Jakob Nikolas Kather. From whole-slide im- age to biomarker prediction: end-to-end weakly supervised deep learning in computational pathology. Nature Protocols, 2024
- MoCo-v3
Xinlei Chen*, Saining Xie*, and Kaiming He. An empirical study of training self-supervised vision transformers. arXiv preprint arXiv:2104.02057, 2021
Citation
If you find our work useful in your research or if you use parts of this code please consider citing our preprint:
@misc{cobra,
title={Unsupervised Foundation Model-Agnostic Slide-Level Representation Learning},
author={Tim Lenz* and Peter Neidlinger* and Marta Ligero and Georg Wölflein and Marko van Treeck and Jakob Nikolas Kather},
year={2024},
eprint={2411.13623},
archivePrefix={arXiv},
primaryClass={cs.CV},
url={https://arxiv.org/abs/2411.13623},
}