U-Net
This repository contains an implementation of U-Net [1]. unet.py implements the class UNet. The implementation has been tested with PyTorch 2.7.1 and CUDA 12.6.
You can load the U-Net from PyTorch Hub.
import torch
# These are the default parameters. They are written out for clarity. Currently no pretrained weights are available.
model = torch.hub.load('sm00thix/unet', 'unet', pretrained=False, in_channels=3, out_channels=1, pad=True, bilinear=True, normalization=None)
# or
# model = torch.hub.load('sm00thix/unet', 'unet_bn', **kwargs) # Convenience function equivalent to torch.hub.load('sm00thix/unet', 'unet', normalization='bn', **kwargs)
# or
# model = torch.hub.load('sm00thix/unet', 'unet_ln', **kwargs) # Convenience function equivalent to torch.hub.load('sm00thix/unet', 'unet', normalization='ln', **kwargs)
# or
# model = torch.hub.load('sm00thix/unet', 'unet_medical', **kwargs) # Convenience function equivalent to torch.hub.load('sm00thix/unet', 'unet', in_channels=1, out_channels=1, **kwargs)
# or
# model = torch.hub.load('sm00thix/unet', 'unet_transconv', **kwargs) # Convenience function equivalent to torch.hub.load('sm00thix/unet', 'unet', bilinear=False, **kwargs)
You can also clone this repository to access the U-Net directly.
import torch
from unet import UNet
model = UNet(in_channels=3, out_channels=1, pad=True, bilinear=True, normalization=None)
Options
The UNet class provides the following options for customization.
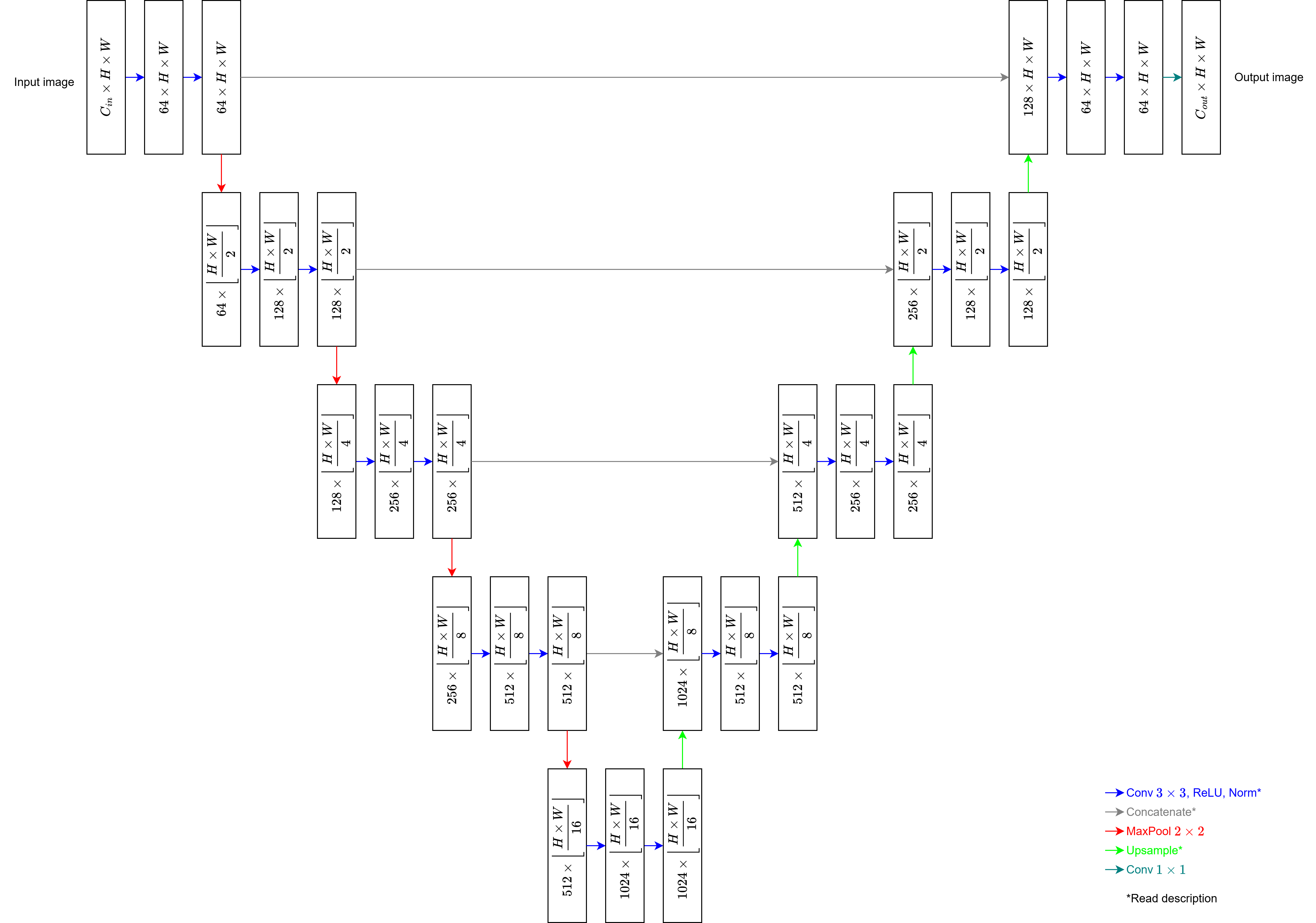
- Number of input and output channels
in_channelsis the number of channels in the input image.out_channelsis the number of channels in the output image. - Upsampling
bilinear = False: Transposed convolution with a 2x2 kernel applied with stride 2. This is followed by a ReLU.bilinear = True: Factor 2 bilinear upsampling followed by convolution with a 1x1 kernel applied with stride 1.
- Padding
pad = True: The input size is retained in the output by zero-padding convolutions and, if necessary, the results of the upsampling operations.pad = False: The output is smaller than the input as in the original implementation. In this case, every 3x3 convolution layer reduces the height and width by 2 pixels each. Consequently, the right side of the U-Net has a smaller spatial size than the left size. Therefore, before concatenating, the central slice of the left tensor is cropped along the spatial dimensions to match those of the right tensor.
- Normalization following the ReLU which follows each convolution and transposed convolution.
normalization = None: Applies no normalization.normalization = "bn": Applies batch normalization [2].normalization = "ln": Applies layer normalization [3]. A permutation of dimensions is performed before the layer to ensure normalization is applied over the channel dimension. Afterward, the dimensions are permuted back to their original order.
In particular, setting bilinear = False, pad = False, and normalization = None will yield the U-Net as originally designed. Generally, however, bilinear = True is recommended to avoid checkerboard artifacts.
As in the original implementation, all weights are initialized by sampling from a Kaiming He Normal Distribution [4], and all biases are initialized to zero. If Batch Normalization or Layer Normalization is used, the weights of those layers are initialized to one and their biases to zero.
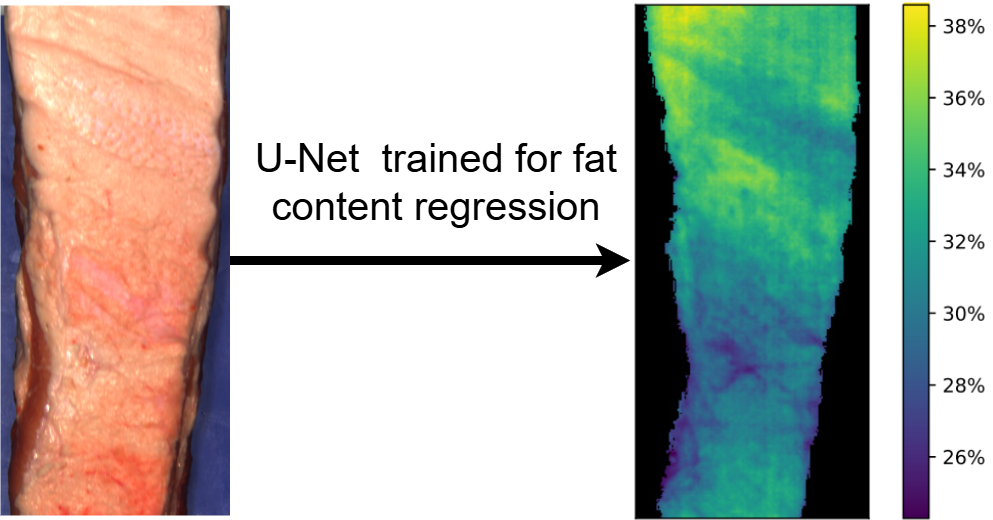
If you use this U-Net implementation, please cite Engstrøm et al. [5] who developed this implementation as part of their work on chemical map geenration of fat content in images of pork bellies.
Citation
If you use the code shared in this repository, please cite this work: https://arxiv.org/abs/2504.14131. The U-Net implementation in this repository was used to generate pixel-wise fat predictions in an image of a pork belly.
References
- O. Ronneberger, P. Fischer, and Thomas Brox (2015). U-Net: Convolutional Networks for Biomedical Image Segmentation. MICCAI 2015.
- S. Ioffe and C. Szegedy (2015). Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. ICML 2015.
- J. L. Ba and J. R. Kiros and G. E. Hinton (2016). Layer Normalization.
- K. He and X. Zhang and S. Ren and J. Sun (2015). Delving Deep into Rectifiers: Surpassing Human-Level Performance on ImageNet Classification.
- O.-C. G. Engstrøm and M. Albano-Gaglio and E. S. Dreier and Y. Bouzembrak and M. Font-i-Furnols and P. Mishra and K. S. Pedersen (2025). Transforming Hyperspectral Images Into Chemical Maps: A Novel End-to-End Deep Learning Approach.
Funding
This work has been carried out as part of an industrial Ph. D. project receiving funding from FOSS Analytical A/S and The Innovation Fund Denmark. Grant number 1044-00108B.