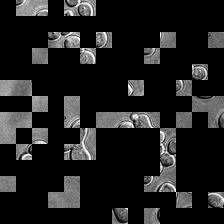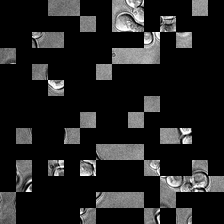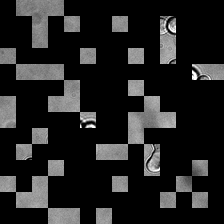import torch
import numpy as np
from PIL import Image
from einops import repeat
from datasets import load_dataset, concatenate_datasets
from IPython.display import display, HTML
from torchvision.transforms import ToPILImage, PILToTensor, Compose
from torchvision.transforms import Resize, RandomCrop, CenterCrop, RandomHorizontalFlip, RandomVerticalFlip, RandomRotation
from vit_pytorch.mae import MAE
from vit_pytorch.simple_vit_with_register_tokens import SimpleViT
from einops.layers.torch import Rearrange
class Args: pass
device = "cpu"
checkpoint = torch.load("v0.0.1.pt",map_location="cpu")
args = checkpoint['args']
args.crops_per_sample = 1
encoder = SimpleViT(
image_size = args.img_dim[1],
channels = args.img_dim[0],
patch_size = args.patch_size,
num_classes = args.num_classes,
dim = args.embed_dim,
depth = args.depth,
heads = args.heads,
mlp_dim = args.mlp_dim,
dim_head = args.embed_dim//args.heads,
).to(device)
model = MAE(
encoder=encoder,
decoder_dim=args.embed_dim,
masking_ratio=args.masking_ratio,
decoder_depth=args.decoder_depth,
decoder_heads=args.heads,
decoder_dim_head=args.embed_dim//args.heads,
).to(device)
model.load_state_dict(checkpoint['model_state_dict'])
<All keys matched successfully>
dataset = load_dataset("danjacobellis/cell_synthetic_labels")
transforms = Compose([
RandomCrop(896),
RandomRotation(22.5),
CenterCrop(672),
Resize(224, interpolation=Image.Resampling.LANCZOS),
RandomVerticalFlip(0.5),
RandomHorizontalFlip(0.5),
PILToTensor(),
])
def collate_fn(batch):
batch_size = len(batch)*args.crops_per_sample
inputs = torch.zeros(
(batch_size, args.img_dim[0], args.img_dim[1], args.img_dim[2]),
dtype=torch.uint8
)
for i_sample, sample in enumerate(batch):
img = sample['image']
for i_crop in range(args.crops_per_sample):
ind = i_sample*args.crops_per_sample + i_crop
inputs[ind,:,:,:] = transforms(img)
return inputs
data_loader_valid = torch.utils.data.DataLoader(
dataset['validation'],
batch_size=8,
shuffle=False,
num_workers=args.num_workers,
drop_last=False,
pin_memory=True,
collate_fn=collate_fn
)
with torch.no_grad():
x = next(iter(data_loader_valid))
x = x.to(torch.float)
x = x / 255
x = x.to(device)
patches = model.to_patch(x)
batch, num_patches, *_ = patches.shape
tokens = model.patch_to_emb(patches)
tokens += model.encoder.pos_embedding.to(device, dtype=tokens.dtype)
num_masked = int(model.masking_ratio * num_patches)
rand_indices = torch.rand(batch, num_patches, device = device).argsort(dim = -1)
masked_indices, unmasked_indices = rand_indices[:, :num_masked], rand_indices[:, num_masked:]
batch_range = torch.arange(batch, device = device)[:, None]
tokens = tokens[batch_range, unmasked_indices]
masked_patches = patches[batch_range, masked_indices]
encoded_tokens = model.encoder.transformer(tokens)
decoder_tokens = model.enc_to_dec(encoded_tokens)
unmasked_decoder_tokens = decoder_tokens + model.decoder_pos_emb(unmasked_indices)
mask_tokens = repeat(model.mask_token, 'd -> b n d', b = batch, n = num_masked)
mask_tokens = mask_tokens + model.decoder_pos_emb(masked_indices)
decoder_tokens = torch.zeros(batch, num_patches, model.decoder_dim, device=device)
decoder_tokens[batch_range, unmasked_indices] = unmasked_decoder_tokens
decoder_tokens[batch_range, masked_indices] = mask_tokens
decoded_tokens = model.decoder(decoder_tokens)
mask_tokens = decoded_tokens[batch_range, masked_indices]
pred_pixel_values = model.to_pixels(mask_tokens)
recon_loss = torch.nn.functional.mse_loss(pred_pixel_values, masked_patches)
def reconstruct_image(self, patches, model_input, masked_indices=None, pred_pixel_values=None, patch_size=8):
patches = patches.cpu()
masked_indices_in = masked_indices is not None
predicted_pixels_in = pred_pixel_values is not None
if masked_indices_in:
masked_indices = masked_indices.cpu()
if predicted_pixels_in:
pred_pixel_values = pred_pixel_values.cpu()
patch_width = patch_height = patch_size
reconstructed_image = patches.clone()
if masked_indices_in or predicted_pixels_in:
for i in range(reconstructed_image.shape[0]):
if masked_indices_in and predicted_pixels_in:
reconstructed_image[i, masked_indices[i].cpu()] = pred_pixel_values[i, :].cpu().float()
elif masked_indices_in:
reconstructed_image[i, masked_indices[i].cpu()] = 0
invert_patch = Rearrange('b (h w) (p1 p2 c) -> b c (h p1) (w p2)', w=int(model_input.shape[3] / patch_width),
h=int(model_input.shape[2] / patch_height), c=model_input.shape[1],
p1=patch_height, p2=patch_width)
reconstructed_image = invert_patch(reconstructed_image)
reconstructed_image = reconstructed_image.numpy().transpose(0, 2, 3, 1)
return reconstructed_image.transpose(0, 3, 1, 2)
with torch.no_grad():
reconstructed_images1 = reconstruct_image(
model,
patches,
x,
masked_indices=masked_indices,
pred_pixel_values=pred_pixel_values,
patch_size=16
)
reconstructed_images2 = reconstruct_image(
model,
patches,
x,
masked_indices=masked_indices,
patch_size=16
)
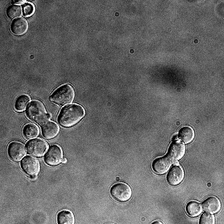
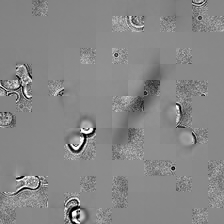
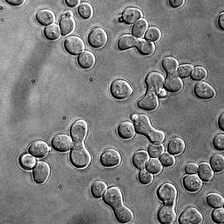
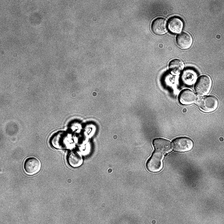
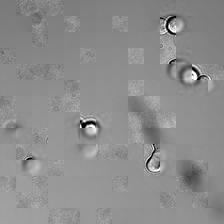
for i_img, img in enumerate(x):
rec1 = reconstructed_images1[i_img]
rec2 = reconstructed_images2[i_img]
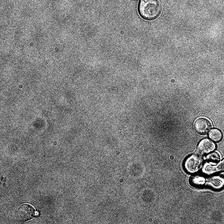
display(ToPILImage()(img[0]))
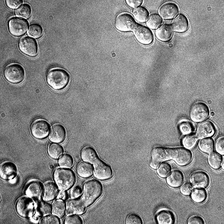
display(ToPILImage()(rec2[0]))
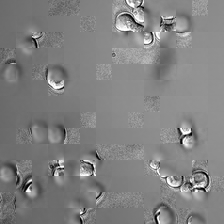
display(ToPILImage()(rec1[0]))
!jupyter nbconvert --to markdown README.ipynb
[NbConvertApp] Converting notebook README.ipynb to markdown
[NbConvertApp] Support files will be in README_files/
[NbConvertApp] Writing 7517 bytes to README.md
Inference Providers
NEW
This model is not currently available via any of the supported Inference Providers.
The model cannot be deployed to the HF Inference API:
The model has no library tag.