Search is not available for this dataset
image
image |
|---|
Introduction
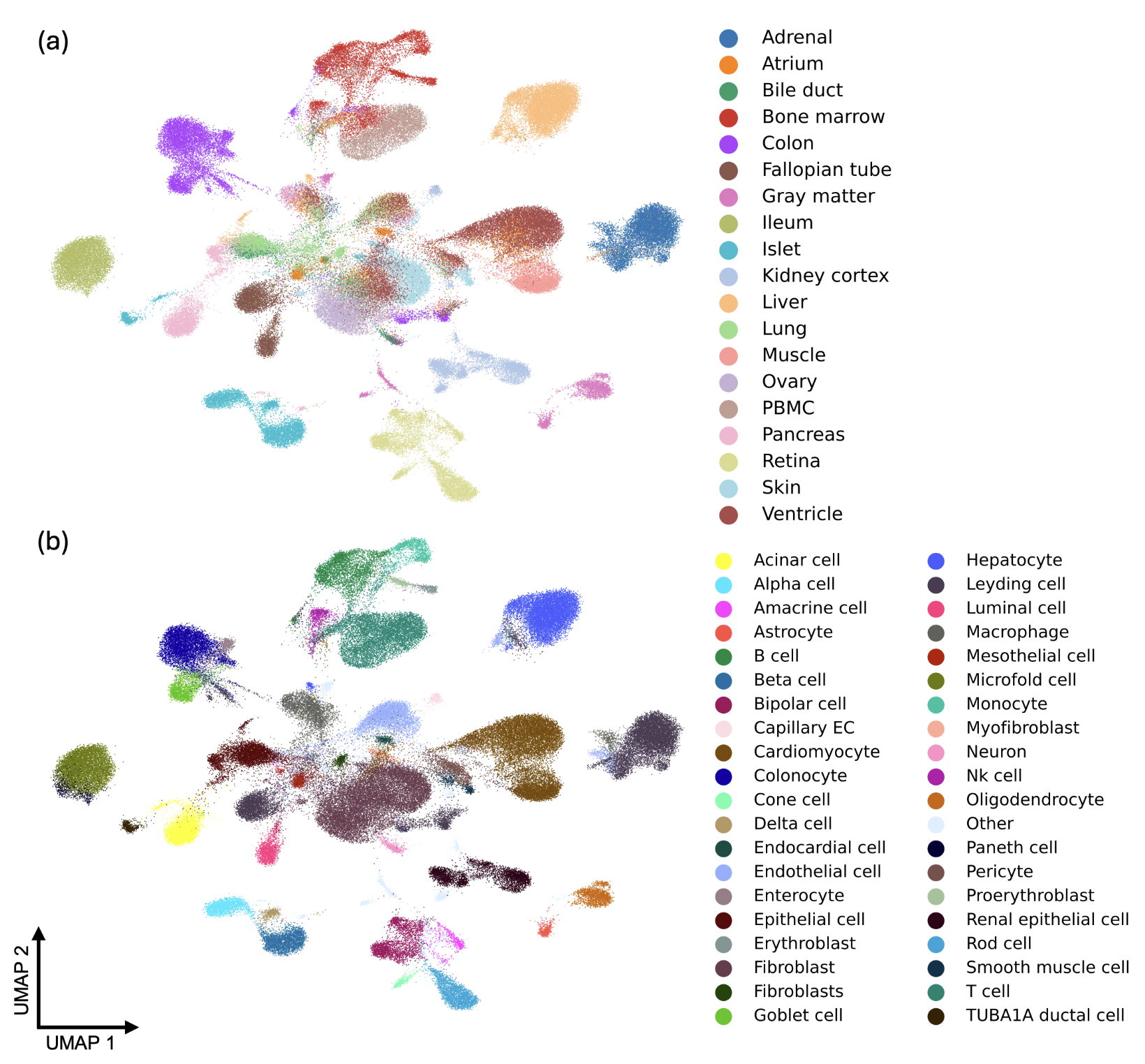
We propose the MiniAtlas dataset, containing more than 100,000 scATAC-seq with paired scRNA-seq as training data, across 19 tissues and 56 cell types, facilitating the training of foundation models. This dataset can be used to training single-cell multiomics fundation model.
Subsets
This dataset is divided into four subsets to accommodate different research needs and access limitations:
full_atlas_atac.h5adandfull_atlas_rna.h5ad(~120k samples): full data of MiniAtlas, containing all tissues and cell types.- Evaluation set for different tissues: containing three tissues (Kidney, PBMC, BMMC), can be used to cell-type annotation or RNA-prediction fine-tuning and evaluation.
Citation
If you find MiniAtlas useful for your research and applications, please cite using this BibTeX:
@article {Wu2025.02.05.636688,
author = {Wu, Juncheng and Wan, Changxin and Ji, Zhicheng and Zhou, Yuyin and Hou, Wenpin},
title = {EpiFoundation: A Foundation Model for Single-Cell ATAC-seq via Peak-to-Gene Alignment},
elocation-id = {2025.02.05.636688},
year = {2025},
doi = {10.1101/2025.02.05.636688},
URL = {https://www.biorxiv.org/content/early/2025/02/08/2025.02.05.636688},
eprint = {https://www.biorxiv.org/content/early/2025/02/08/2025.02.05.636688.full.pdf},
journal = {bioRxiv}
}
- Downloads last month
- 96