viewer: false
license:
- apache-2.0
language:
- en
library_name:
- transformers
What is it?
Data quantity and quality play a vital role in determining the performance of Large Language Models (LLMs). High-quality data, in particular, can significantly boost the LLM’s ability to generalize on a wide range of downstream tasks. To cater to the requirements of the Granite models, we focused on a goal to produce a 10T dataset, named, GneissWeb, that is of higher quality than all other datasets of similar size available. Gneiss, pronounced as nice, is a strong and durable rock that is used for building and construction.
The GneissWeb recipe consists of sharded exact substring deduplication and a judiciously constructed ensemble of quality filters. We present the key evaluations that guided our design choices and provide filtering thresholds that can be used to filter the dataset to match the token and quality needs of Stage-1 (early pre-training) or Stage-2 (annealing) datasets.
Our evaluations demonstrate that GneissWeb outperforms state-of-the-art large open datasets (5T+ tokens). Specifically, ablation models trained on GneissWeb outperform those trained on FineWeb.V1.1.0 by 2.14 percentage points in terms of average score computed on a set of 11 benchmarks (both zero-shot and few-shot) commonly used to evaluate pre-train datasets. When the evaluation set is extended to 20 benchmarks (both zero-shot and few-shot), ablation models trained on GneissWeb outperform those trained on FineWeb.V1.1.0 by 1.49 percentage points.
For more details about GneissWeb are available in this blog and this paper.
The GneissWeb Recipe in a Nutshell : Building on Top of FineWeb
Hugging Face introduced FineWeb V1.1.0, a large-scale dataset for LLM pre-training, consisting of 15 trillion tokens (44TB disk space). We started with the goal of distilling 10T+ high quality tokens from FineWeb V1.1.0, so that we get sufficiently large number of quality tokens suitable for Stage-1 pre-training. Unlike the FineWeb.Edu families, which rely on a single quality annotator and perform aggressive filtering, we developed a multi-faceted ensemble of quality annotators to enable fine-grained quality filtering. This allowed us to achieve a finer trade-off between the quality and quantity of the tokens retained. While the GneissWeb recipe is focused at obtaining 10T+ high quality tokens suitable for Stage-1 pre-training, it is also possible to adapt the recipe by tuning filtering parameters to produce smaller and higher quality datasets fit for Stage-2 kind of training.
An Overview of the GneissWeb Recipe
The GneissWeb dataset was obtained by applying the following processing steps to Fineweb :
- Exact substring deduplication at line level
- Custom built Fasttext quality filter
- Custom built Fasttext category classifier
- Custom built Category-aware readability score quality filter
- Custom built Category-aware extreme_tokenized quality filter
These were applied in the order shown in the Fig 1
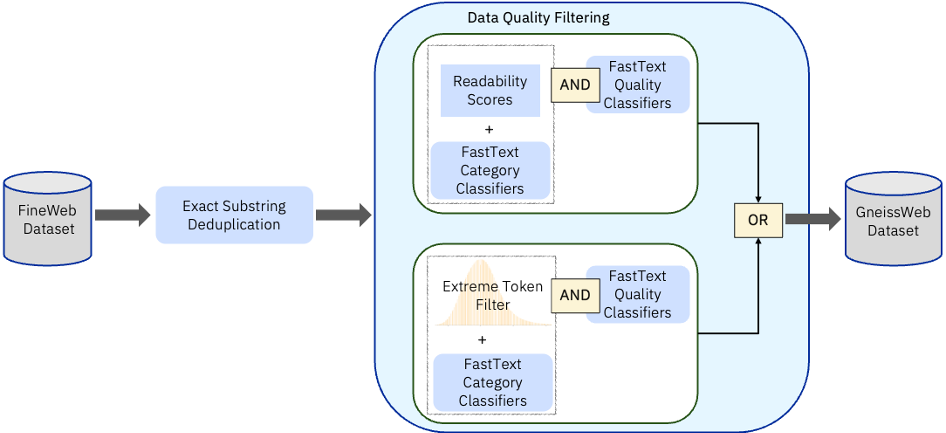
Figure 1 : GneissWeb recipe
The net impact was that the dataset size of 15T tokens was filtered down to approx 10T tokens. In subsequent sections we describe the overall performance obtained using GneissWeb compared to other baselines.
Evaluation Strategy
To compare GneissWeb against the baselines, we trained decoder models of 7B parameters on a Llama architecture. These were trained on 350B tokens to validate the performance of each processing step. The data was tokenized using a starcoder tokenizer and training was done with a sequence length of 8192.
We used FineWeb 1.1.0 and FineWeb-Edu-score2 as our comparison baselines. The former is of 15T tokens and the latter of 5.4T tokens. Fig 2 shows how the subsamples were created for the Fineweb baselines as well for GneissWeb.
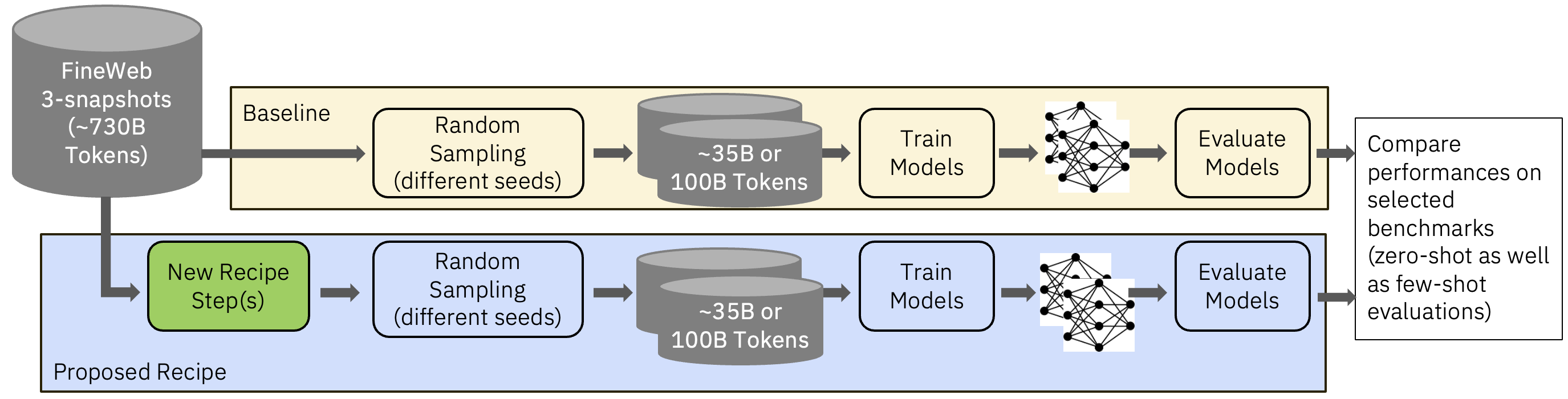
Figure 2: Subsampling and Ablation Strategy
Evaluation Benchmarks Selection
We evaluated our ablation models using lm-evaluation-harness on two categories of tasks: 11 High-Signal tasks (0-shot and few-shot) and 20 Extended tasks (0-shot and few-shot).
High-Signal tasks:
Similar to FineWeb, we used the following criteria for selecting the 11 High-Signal/Early-Signal tasks: accuracy above random guessing, accuracy monotonically increasing over training epochs, and small variance across runs. These are shown in Fig 3 and cover Commonsense Reasoning, Reading Comprehension, World Knowledge and Language Understanding task categories. We used both the zero-shot as well as few-shot variations of these tasks.
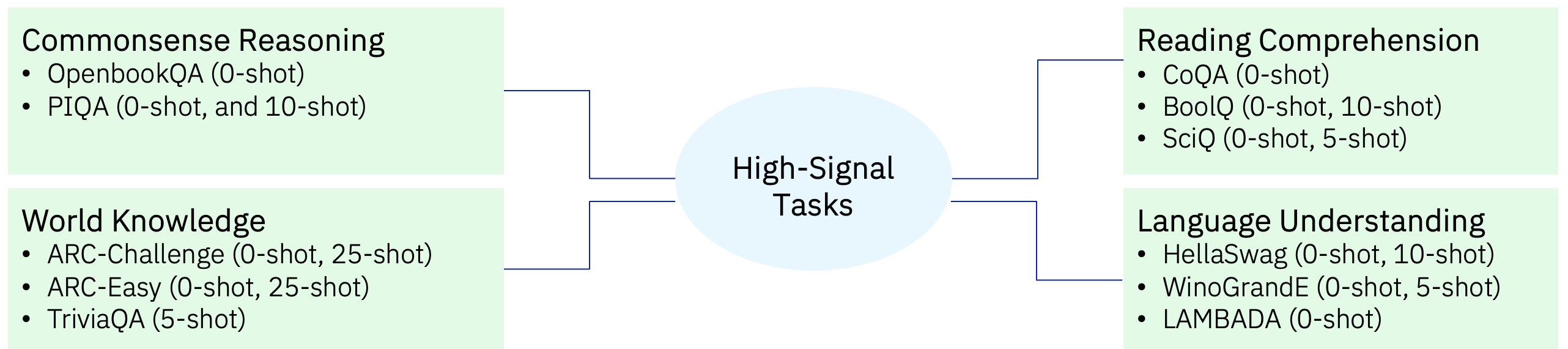
Figure 3: High Signal Tasks — provide good signal at relatively small scale (of 1.4B models trained on 35B to 100B tokens)
The High-Signal tasks were used to analyze individual ingredients and possible recipe combinations via ablations. After we narrowed a few candidate recipes using these signals, we used the extended set of benchmarks to evaluate the model’s ability to generalize.
Extended tasks:
The extended tasks shown in Fig 4 are a superset of the High Signal tasks. Besides the task categories of Commonsense Reasoning, Reading Comprehension, World Knowledge, Language Understanding, it also has the category of Symbolic Problem Solving. For the extended set, we also focus on zero-shot as well as few-shot variations.
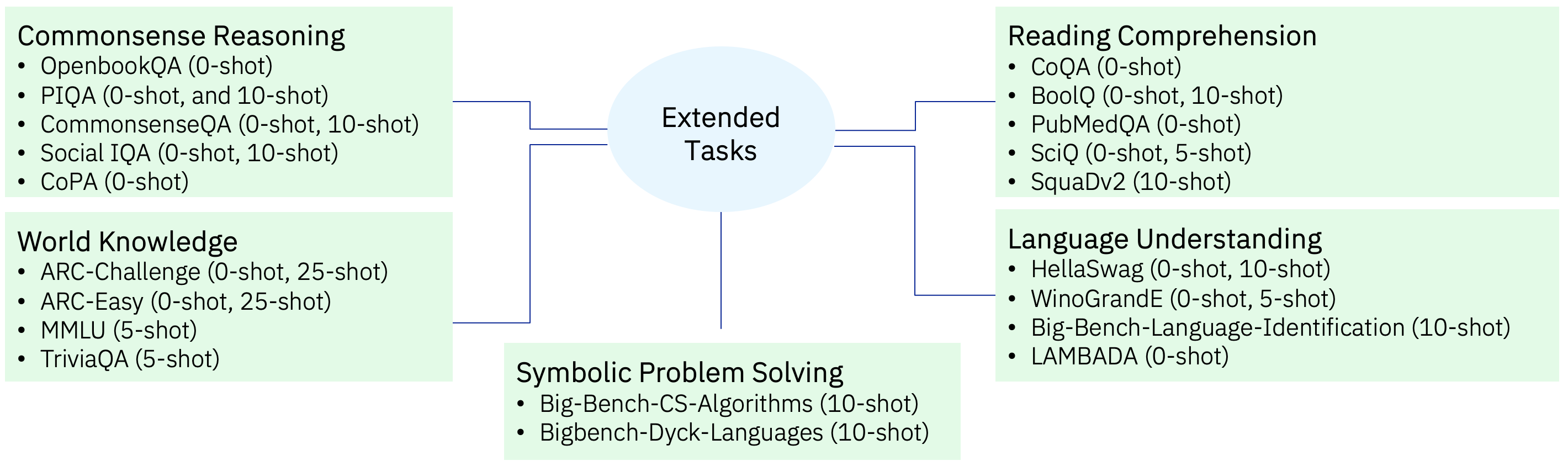
Figure 4: Extended Tasks — broader set of tasks to evaluate generalization at larger number of tokens and/or larger model sizes
The Extended Task set have some tasks which are not in High Signal. These tasks are useful but at ablation scale may have high standard deviation (like PubMedQA) or are at random guessing the entire training cycle (like MMLU) or which are above random guessing but do not show improvement with training (like GSM8k). However, these tasks are useful indicators for larger model performance and thus have been retained in the Extended Tasks set.
Evaluation Results at 7 Billion Model Size with 350 Billion Tokens
Given than training models of size 7 Billion parameters require lot more compute and so does evaluation, we have limited training to 350 billion tokens. We see that the models trained on GneissWeb outperform the models trained on FineWeb.V1.1.0 and FineWeb-Edu-score-2.

Figure 5: Comparison of Average Eval Scores on High Signal and Extended Eval Tasks at 7B model size. Scores are averaged over 3 random seeds used for data sampling and are reported along with standard deviations.
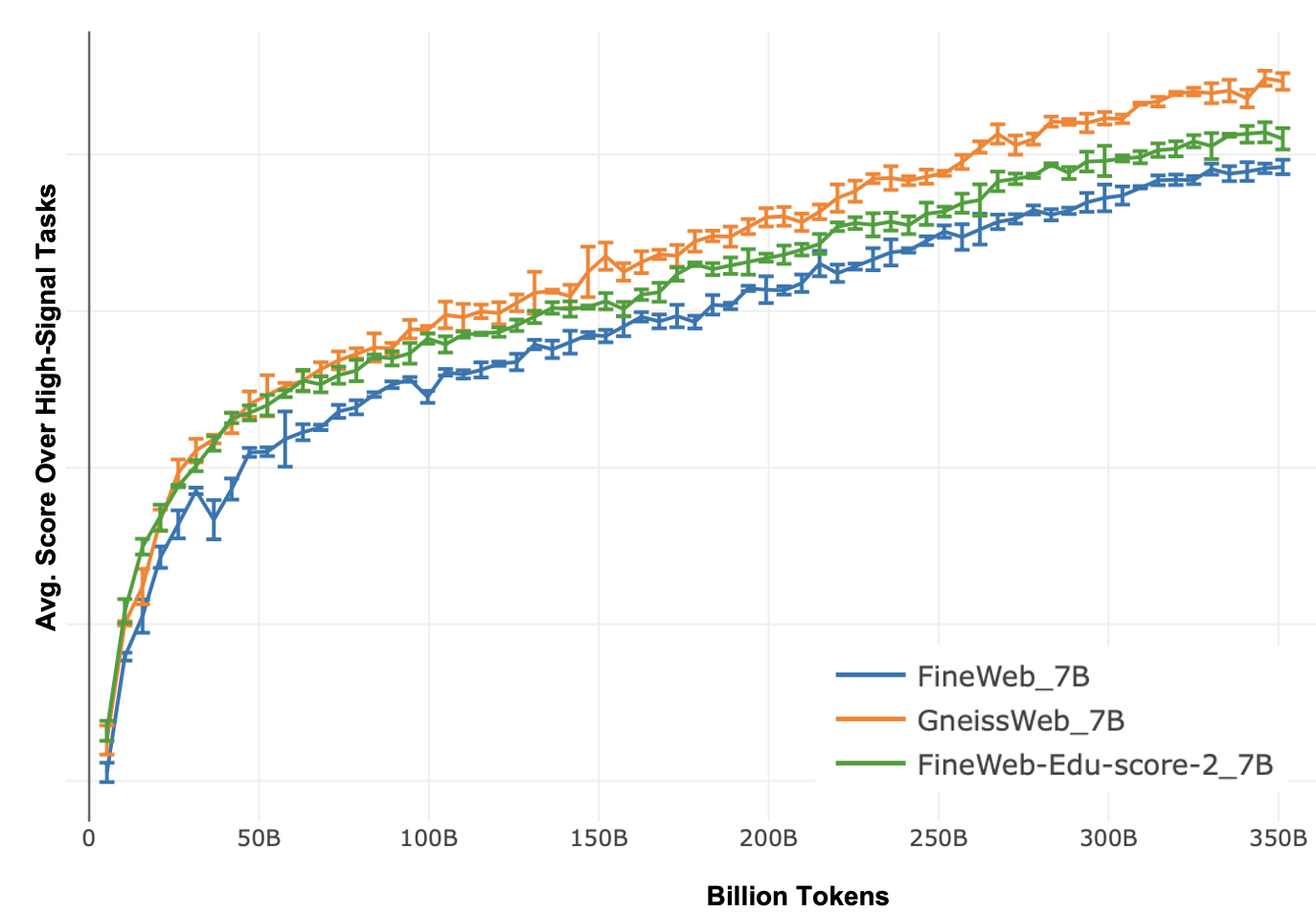
Figure 6: Average evaluation score on High-Signal tasks versus the number of tokens at 7 Billion model size for 350 Billion tokens. The model trained on GneissWeb consistently outperforms the one trained on FineWeb.V1.1 throughout the training.
Dataset Summary
Recently, IBM has introduced GneissWeb; a large dataset yielding around 10 trillion tokens that caters to the data quality and quantity requirements of training LLMs. The models trained using GneissWeb dataset outperform those trained on FineWeb 1.1.0 by 2.14 percentage points in terms of average score computed on a set of 11 commonly used benchmarks
Developers: IBM Research
Release Date: Feb 10th, 2025
License: Apache 2.0.
What is being released?
fastText models used in the curation of GneissWeb
-
The fastText model takes as input text and classifies whether the text is ''high-quality'' (labeled as __label__hq) or ''low-quality'' (labeled as __label__cc). The model can be used with python (please refer to fasttext documentation for details on using fasttext classifiers) or with IBM Data Prep Kit (DPK) (please refer to the example notebook for using a fastText model with DPK). The GneissWeb ensemble filter uses the confidence score given to __label__hq for filtering documents based on an appropriately chosen threshold. The fastText model is used along with [DCLM-fastText] (https://huggingface.co/mlfoundations/fasttext-oh-eli5) and other quality annotators.
Classifiers for Science, Technology, Medical and Education
There are separate models for Science, Technology, Medical and Education. Each fastText model takes as input a text and classifies whether the text categorized as for its subject (labeled as
__label__hq) or other categories''cc'' (labeled as__label__cc). The model can be used with python (please refer to fasttext documentation for details on using fasttext classifiers) or with IBM Data Prep Kit (DPK) (please refer to the example notebook for using a fastText model with DPK).The GneissWeb ensemble filter uses the confidence score given to
__label__hqfor filtering documents based on an appropriately chosen threshold. The fastText models are used together along with other quality annotators.
-
Bloom filter built on the document ids of GneissWeb documents. This can be used to recreat GneissWeb using the document ids from FineWeb 1.1.0 or any version of Common Crawl
This filter offers a way to determine which documents of FineWeb 1.1.0 or Common Crawl are part of GneissWeb. Bloom Annotatory transforms are available in IBMs Data Prep Kit to make it easy to use this filter.
The Bloom Annotator transform assigns a label of 1 if the document is present in the GneissWeb Bloom filter; otherwise, it assigns 0. This approach provides a clear understanding of which documents in FineWeb 1.1.0 are also present in GneissWeb and which are not.
The id column in FineWeb 1.1.0 looks like this : urn:uuid:39147604-bfbe-4ed5-b19c-54105f8ae8a7. The bloom filter is of the rbloom type and of size 28 GB.
IBM Data Prep Kit transforms and notebook to recreate GneissWeb using the methods described above
Notebook to recreate GneissWeb using a bloom filter built on the document ids of GneissWeb