Acknowledge license to accept the repository
By agreeing, you accept to share your contact information (email and username) with the repository authors and confirm that you will not use the dataset for harmful, unethical, or malicious purposes. By agreeing, you confirm that you will not distribute, or share any part of the dataset. If another user within your organization wishes to access the dataset, they must register as an individual user and agree to comply with the terms of use. Please allow 3-5 business days for our team to process your request.
By agreeing, you accept to share your contact information (email and username) with the repository authors.
Log in or Sign Up to review the conditions and access this dataset content.
Dataset card for TCGA digital spatial transcriptomics data
This repository contains results from the paper "DeepSpot: Leveraging Spatial Context for Enhanced Spatial Transcriptomics Prediction from H&E Images".
Authors: Kalin Nonchev, Sebastian Dawo, Karina Selina, Holger Moch, Sonali Andani, Tumor Profiler Consortium, Viktor Hendrik Koelzer, and Gunnar Rätsch
The preprint is available here.
What is TCGA digital spatial transcriptomic?
We trained a model using available spatial transcriptomics data to predict gene expression for both fresh frozen (FF) and formalin-fixed paraffin-embedded (FFPE) slides from TCGA SKCM (skin melanoma) and TCGA KIRC (kidney cancer) datasets. More information can be found at: https://github.com/ratschlab/DeepSpot.
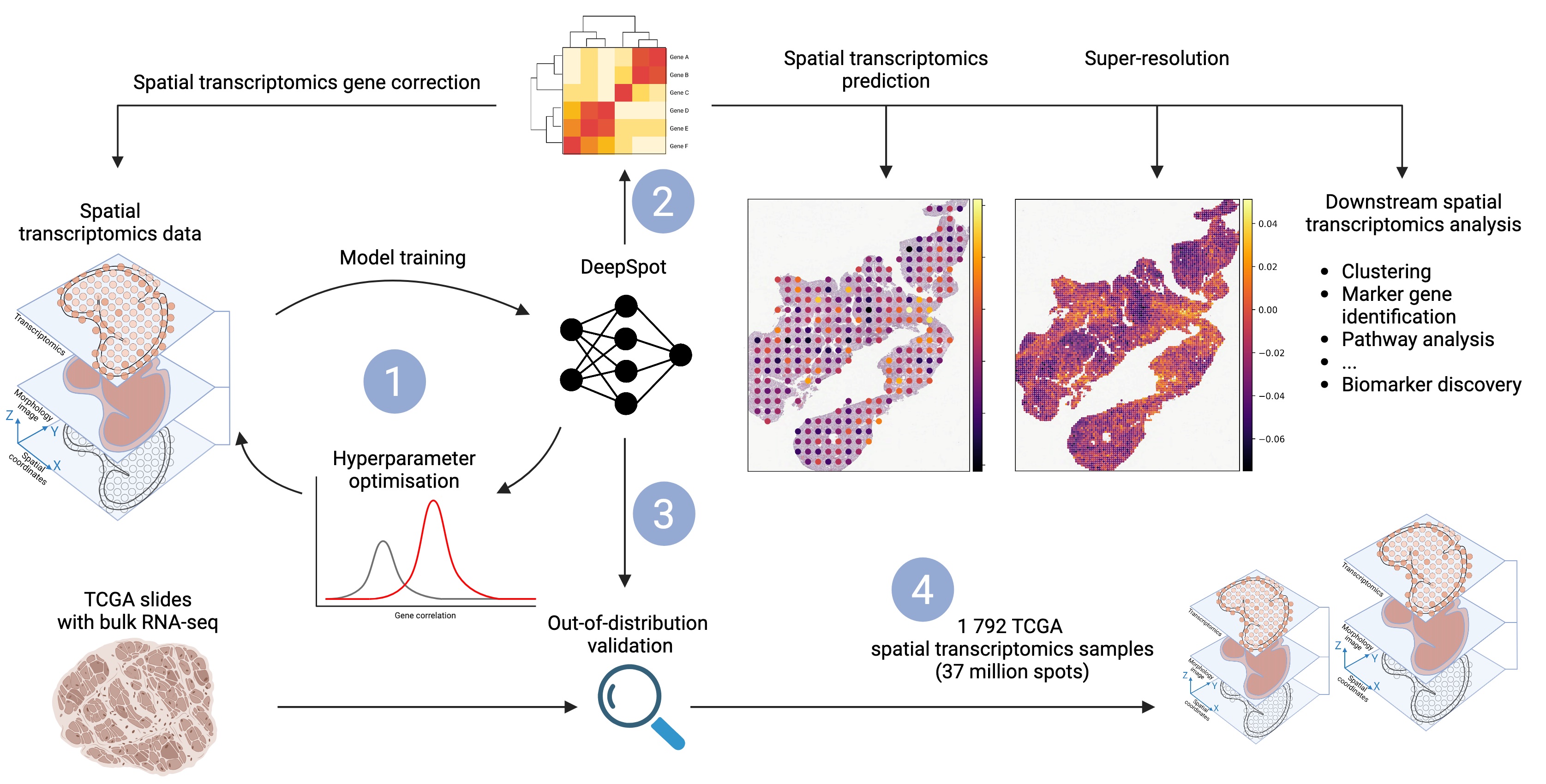 Fig: DeepSpot predicts spatial transcriptomics from H&E images by leveraging recent foundation models in pathology and spatial multi-level tissue context. 1: DeepSpot is trained to predict 5 000 genes, with hyperparameters optimized using cross-validation. 2: DeepSpot
can be used for de novo spatial transcriptomics prediction or for correcting existing spatial transcriptomics data. 3: Validation involves nested leave-one-out patient cross-validation and
out-of-distribution testing. We predicted spatial transcriptomics from TCGA slide images, aggregated the data into pseudo-bulk RNA profiles, and compared them with the available
ground truth bulk RNA-seq. 4: DeepSpot generated 1 792 TCGA spatial transcriptomics samples with over 37 million spots from melanoma or kidney cancer patients, enriching the
available spatial transcriptomics data for TCGA samples and providing valuable insights into the molecular landscapes of cancer tissues.
Fig: DeepSpot predicts spatial transcriptomics from H&E images by leveraging recent foundation models in pathology and spatial multi-level tissue context. 1: DeepSpot is trained to predict 5 000 genes, with hyperparameters optimized using cross-validation. 2: DeepSpot
can be used for de novo spatial transcriptomics prediction or for correcting existing spatial transcriptomics data. 3: Validation involves nested leave-one-out patient cross-validation and
out-of-distribution testing. We predicted spatial transcriptomics from TCGA slide images, aggregated the data into pseudo-bulk RNA profiles, and compared them with the available
ground truth bulk RNA-seq. 4: DeepSpot generated 1 792 TCGA spatial transcriptomics samples with over 37 million spots from melanoma or kidney cancer patients, enriching the
available spatial transcriptomics data for TCGA samples and providing valuable insights into the molecular landscapes of cancer tissues.
The data includes spatial transcriptomics for:
- TCGA SKCM
- 472 FF slides
- 276 FFPE slides
- TCGA KIRC
- 528 FF slides
- 516 FFPE slides
Folder tree:
HF
├── TCGA_KIRC
│ ├── FF
│ └── FFPE
└── TCGA_SKCM
├── FF
└── FFPE
How to start?
pip install datasets
Logging
from huggingface_hub import login, hf_hub_download, snapshot_download
import squidpy as sq
import pandas as pd
import scanpy as sc
import datasets
login(token="YOUR HUGGINGFACE TOKEN")
Load metadata information
# Define dataset details
repo_id = "nonchev/TCGA_digital_spatial_transcriptomics"
filename = "metadata_2025-01-11.csv"
# Create path
file_path = hf_hub_download(repo_id=repo_id, filename=filename, repo_type="dataset")
# Load metata
metadata = pd.read_csv(file_path)
metadata.head()
dataset slide_type sample_id n_spots file_path
0 TCGA_SKCM FFPE TCGA-BF-AAP6-01Z-00-DX1.EFF1D6E1-CDBC-4401-A10... 5860 TCGA_SKCM/FFPE/TCGA-BF-AAP6-01Z-00-DX1.EFF1D6E...
1 TCGA_SKCM FFPE TCGA-FS-A1ZU-06Z-00-DX3.0C477EE6-C085-42BE-8BA... 2856 TCGA_SKCM/FFPE/TCGA-FS-A1ZU-06Z-00-DX3.0C477EE...
2 TCGA_SKCM FFPE TCGA-D9-A1X3-06Z-00-DX1.17AC16CC-5B22-46B3-B9C... 6236 TCGA_SKCM/FFPE/TCGA-D9-A1X3-06Z-00-DX1.17AC16C...
Download a single TCGA spatial transcriptomics sample
local_dir = 'TCGA_data' # Change the folder path as needed
snapshot_download("nonchev/TCGA_digital_spatial_transcriptomics",
local_dir=local_dir,
allow_patterns="TCGA_SKCM/FFPE/TCGA-D9-A3Z3-06Z-00-DX1.C4820632-C64D-4661-94DD-9F27F75519C3.h5ad.gz",
repo_type="dataset")
adata = sc.read_h5ad("path/to/h5ad.gz")
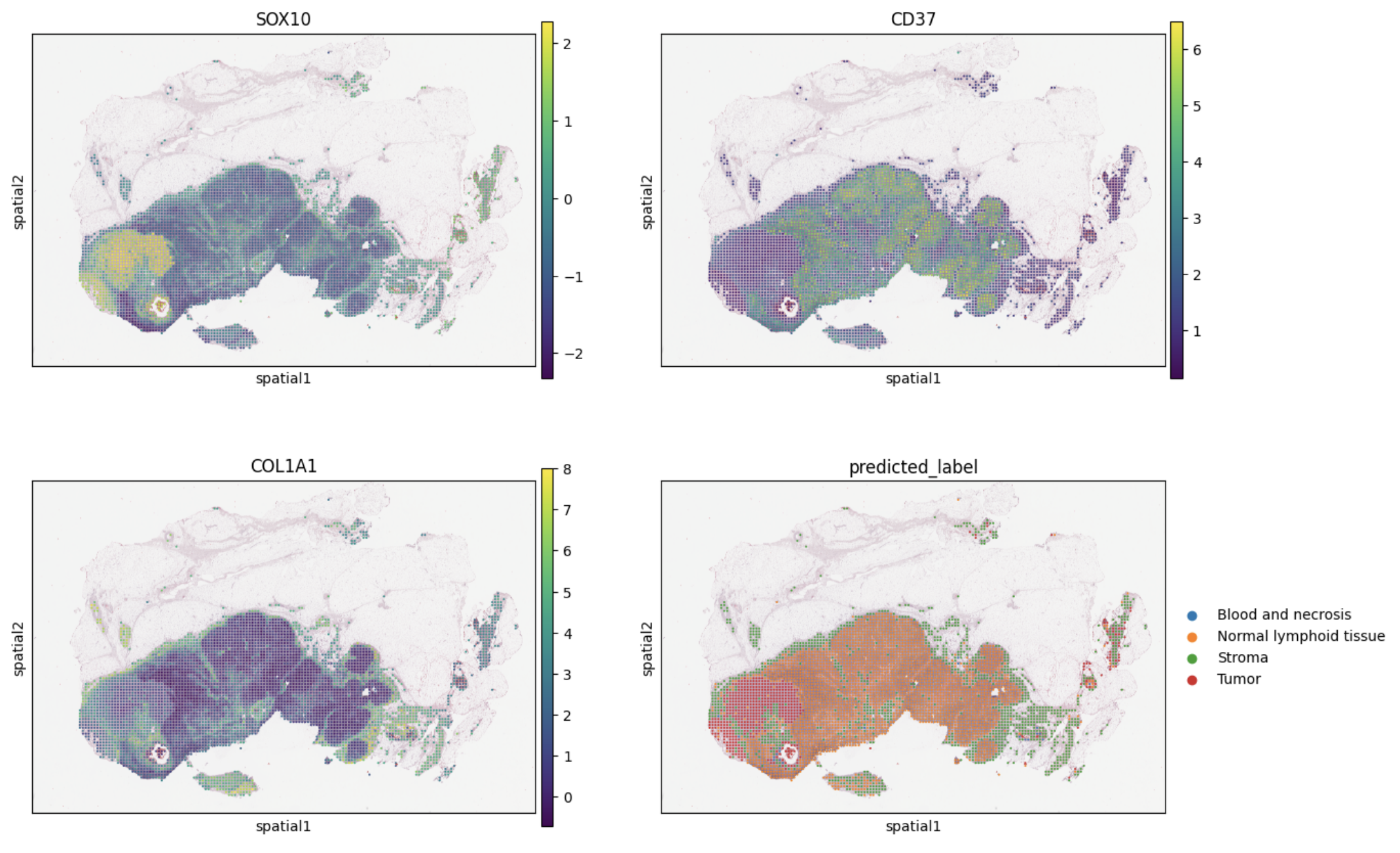
sq.pl.spatial_scatter(adata,
color=["SOX10", "CD37", "COL1A1", "predicted_label"],
size=20, img_alpha=0.8, ncols=2)
Download the entire TCGA digital spatial transcriptomics data
local_dir = 'TCGA_data' # Change the folder path as needed
# Note that the full dataset is around 2TB
snapshot_download("nonchev/TCGA_digital_spatial_transcriptomics",
local_dir=local_dir,
repo_type="dataset")
Download subset of TCGA digital spatial transcriptomics:
import datasets
local_dir='TCGA_data' # will be downloaded to this folder
cancer_type = ['TCGA_KIRC/*'] # OR TCGA_SKCM
## or ['TCGA_KIRC/FF/*'] or ['TCGA_KIRC/FFPE/*'] or based on slide type ["*/FFPE/*", "*/FF/*"]
snapshot_download("nonchev/TCGA_digital_spatial_transcriptomics",
local_dir=local_dir,
allow_patterns=cancer_type,
repo_type="dataset")
Data organization
Each file is of the form {slide_id}.h5ad.gz and can be loaded as:
import scanpy as sc
# Load the data
adata = sc.read_h5ad("../path/to/slide_id.h5ad.gz")
# Note: Since the data is compressed, loading it may take more time.
# It is recommended to uncompress the data if sufficient storage is available.
adata
AnnData object with n_obs × n_vars = 4428 × 5000
obs: 'x_array', 'y_array', 'x_pixel', 'y_pixel', 'barcode', 'predicted_label'
uns: '20x_slide', 'scaled_slide_info', 'spatial'
obsm: 'spatial'
where:
.obs
- x_array and y_array represent the spot coordinates on the image grid.
- x_pixel and y_pixel correspond to the center spot coordinates on the slide, scaled to 20x magnification.
- predicted_label is the label transferred from the training data, obtained by fitting a random forest model on the provided manual annotations for Melanoma or the cluster labels for Kidney Cancer.
- the remaining columns with label names represent the probabilities of the spot being assigned to the predicted_label.
.var
- 20x_slide - original slide image downloaded from TCGA and scaled to 20x magnification
- scaled_slide_info metadata
- spatial - metadata required for squidpy.pl.spatial_scatter
NB: To distinguish in-tissue spots from the background, tiles with a mean RGB value above 200 (near white) were discarded. Additional preprocessing can remove potential image artifacts.
How to cite:
@article{nonchev2025deepspot,
title={DeepSpot: Leveraging Spatial Context for Enhanced Spatial Transcriptomics Prediction from H\&E Images},
author={Nonchev, Kalin and Dawo, Sebastian and Silina, Karina and Moch, Holger and Andani, Sonali and Tumor Profiler Consortium and Koelzer, Viktor H and Raetsch, Gunnar},
journal={medRxiv},
pages={2025--02},
year={2025},
publisher={Cold Spring Harbor Laboratory Press}
}
NB: Computational data analysis was performed at Leonhard Med (https://sis.id.ethz.ch/services/sensitiveresearchdata/) secure trusted research environment at ETH Zurich. Our pipeline aligns with the specific cluster requirements and resources.
- Downloads last month
- 61